Cell Polarization in C.elegans
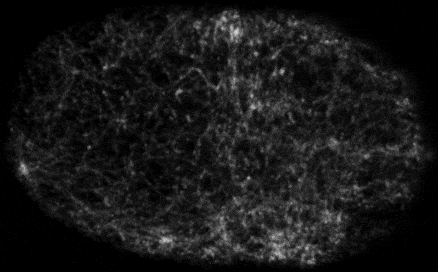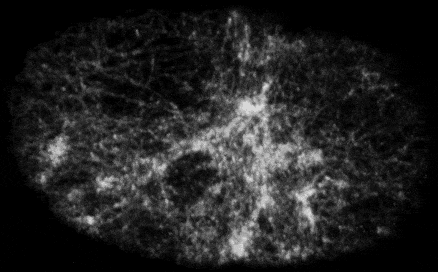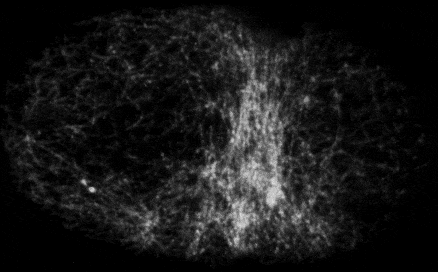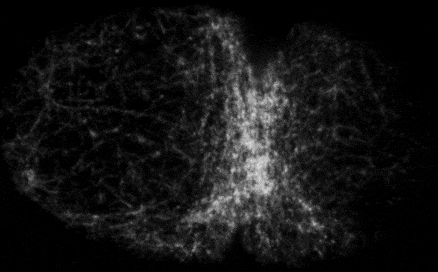
We use the early C. elegans embryo as a model system to understand how cells form and dynamically stabilize cell polarity. In the one-cell C. elegans zygote, polarity is encoded by asymmetric distributions of conserved polarity determinants known as the PAR proteins. These asymmetries are established in response to a transient sperm-derived cue and then maintained by a complex network of interactions among the PAR proteins, small GTPases, and a dynamic actomyosin cytoskeleton. We combine molecular genetics with powerful new microscopy tools (e.g. single molecule imaging and particle tracking analysis) and mathematical modeling to identify the core design principles that govern this fundamental proces
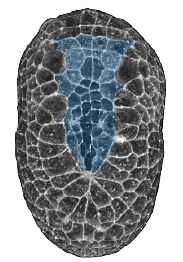
Tissue Morphogenesis in Ascidians
We use Ascidian (sea squirt) embryos to study how embryos pattern force generation in space and time to orchestrate tissue morphogenesis. Ascidian embryos shape themselves using classical morphogenetic mechanisms like invagination, convergent extension and zippering/fusion of epithelial sheets. They do this very fast, with very few cells, under the control of well-characterized regulatory gene networks and highly conserved signaling pathways. Small, optically clear embryos, and well-developed tools for genetic perturbation and transgenesis, provide a unique opportunity to study the dynamic integration of cell fate determination, tissue polarity, and the cytomechanics of tissue morphogenesis. In previous work we have studied dynamics of endoderm invagination during gastrulation, and the dynamics of notochord invagination and elongation. Our current focus is on the dynamics of neural tube closure.
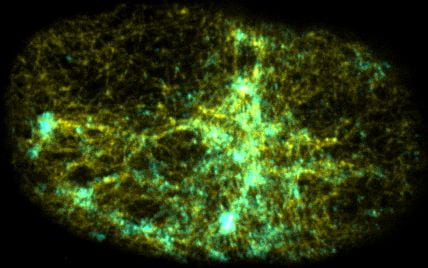
Self-organized actomyosin contractility
Embryonic cells harness actomyosin contractility to move, change shape and to produce long-range flows that redistribute their internal components. We are interested in the basic principles that underlie dynamic control of cortical actomyosin contractility. How do contractility and cortical flow emerge from microscopic interactions among actin filaments, crosslinking proteins, and myosin motors? How do embryonic cells tune contractility through local modulation of filament assembly, disassembly and motor activity? How do they integrate biochemical signaling and contractility to pattern force generation in space and time? We currently focus in three areas: (a) Self-organization of the contractile ring during cytokinesis, (b) Spatiotemporal control of pulsed actomyosin contractility through dynamic coupling of RhoA signaling and actomyosin dynamics. (c) Dynamic coupling of actomyosin contractility and Par protein dynamics during polarization.
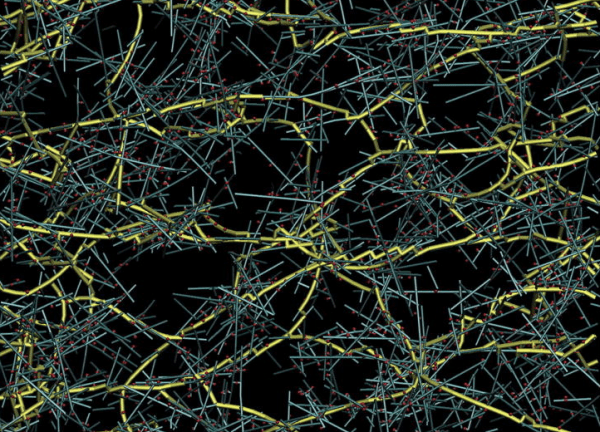
Models for actomyosin contractility and cortical flow
We are using agent-based models to explore how microscopic interactions among actin filaments, crosslinking proteins and myosin motors give rise to macroscopic dynamics of contractility and cortical flow, and how the microscopic properties can be tuned to produce functional variation in the macroscopic dynamics, as observed in living cells or in vitro experiments with purified proteins. Our recent efforts have focused on understanding how the tunable properties of myosin motors shape force buildup and transmission in contractile networks and how actin turnover allows contractile networks continuously build and dissipate force to drive rapid network remodeling and cortical flow.